| TAPREG HELP | |
|
|
|
| TAPREG HELP | |
|
|
|
The transporter associated with antigen processing (TAP) binds and shuttles peptides ranging from 8 to 16 residues from the cytosol to the endoplasmic reticulum, determining the repertoire CD8 T cell epitopes, and TAPREG is the only available server that can predict the binding affinity to TAP of peptides of this variable length.
TAPREG admits two different types of input queries, a protein sequence or one or several peptide sequences, which are selected by users, prior to entering their sequences. Sequences entered in the server must be in FASTA format. Next follows an example of these two types of input.
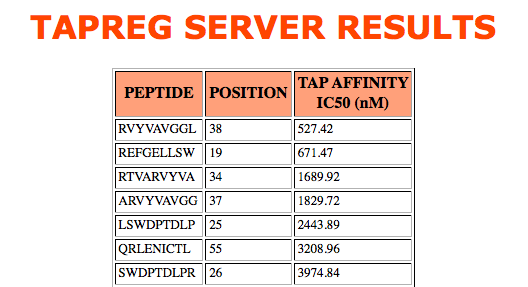
>HLAF_HUMAN MAPRSLLLLLSGALALTDTWAGSHSLRYFSTAVSRPGRGEPRYIAVEYVDDTQFLRFDSDAAIPRMEPREPWVEQEGPQYWEWTTGYA KANAQTDRVALRNLLRRYNQSEAGSHTLQGMNGCDMGPDGRLLRGYHQHAYDGKDYISLNEDLRSWTAADTVAQITQRFYEAEEYAEE FRTYLEGECLELLRRYLENGKETLQRADPPKAHVAHHPISDHEATLRCWALGFYPAEITLTWQRDGEEQTQDTELVETRPAGDGTFQK WAAVVVPSGEEQRYTCHVQHEGLPQPLILRWEQSPQPTIPIVGIVAGLVVLGAVVTGAVVAAVMWRKKSSDRNRGSYSQAAVTDSAQG SGVSLTANKV
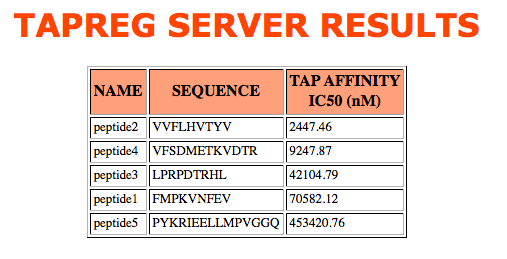
>peptide1 FMPKVNFEV >peptide2 VVFLHVTYV >peptide3 LPRPDTRHL >peptide4 VFSDMETKVDTR >peptide5 PYKRIEELLMPVGGQInput query (Protein or peptides) can be pasted or uploaded from a local file. If the input is uploaded from a local file, users need first to browse/choose the local file and then hit the upload bottom. This is done to facilitatepreprocessing and error checking of the data prior to submission to the server.
The two models available in TAPREG are based on support vector machine regression and were trained on sparse (DS_613 sparse model) and blosum (DS_613 blosum model) representations of 613 9mer peptides of known binding affinity to TAP. Models were only trained in peptide residues 1, 2, 3 and 9, which have been shown to have the major contribution to TAP binding The binding binding affinity of any peptide randing from 8 to 16 is computed by applying the models to peptide fragments encompassing the noted residue selections. The server will return an error for input peptides out of that range.
In cross-validation, the model trained on blosum-encoded sequences achieved a person correlation coefficient (Rp) of
0.83 ± 0.03 while the sparse counterpart achieved an Rp = 0.81 ± 0.03. The correlation achieved by the DS_613 blosum and sparse models
predicting the known TAP binding affinity of a set 98 peptides with a length ranging from 10 to 16 residues (independent test set)
was 0.78 and 0.73, respectively.
Training and testing datasets available upon request from Pedro Reche
| CONTACT: For any questions: Pedro Reche |